Making Changes
Make a change to the repository
For this exercise each participant will create a single new file, setting a few parameters to values of their choice, in their fork. We will then collate everyone’s files in the original repository through pull requests.
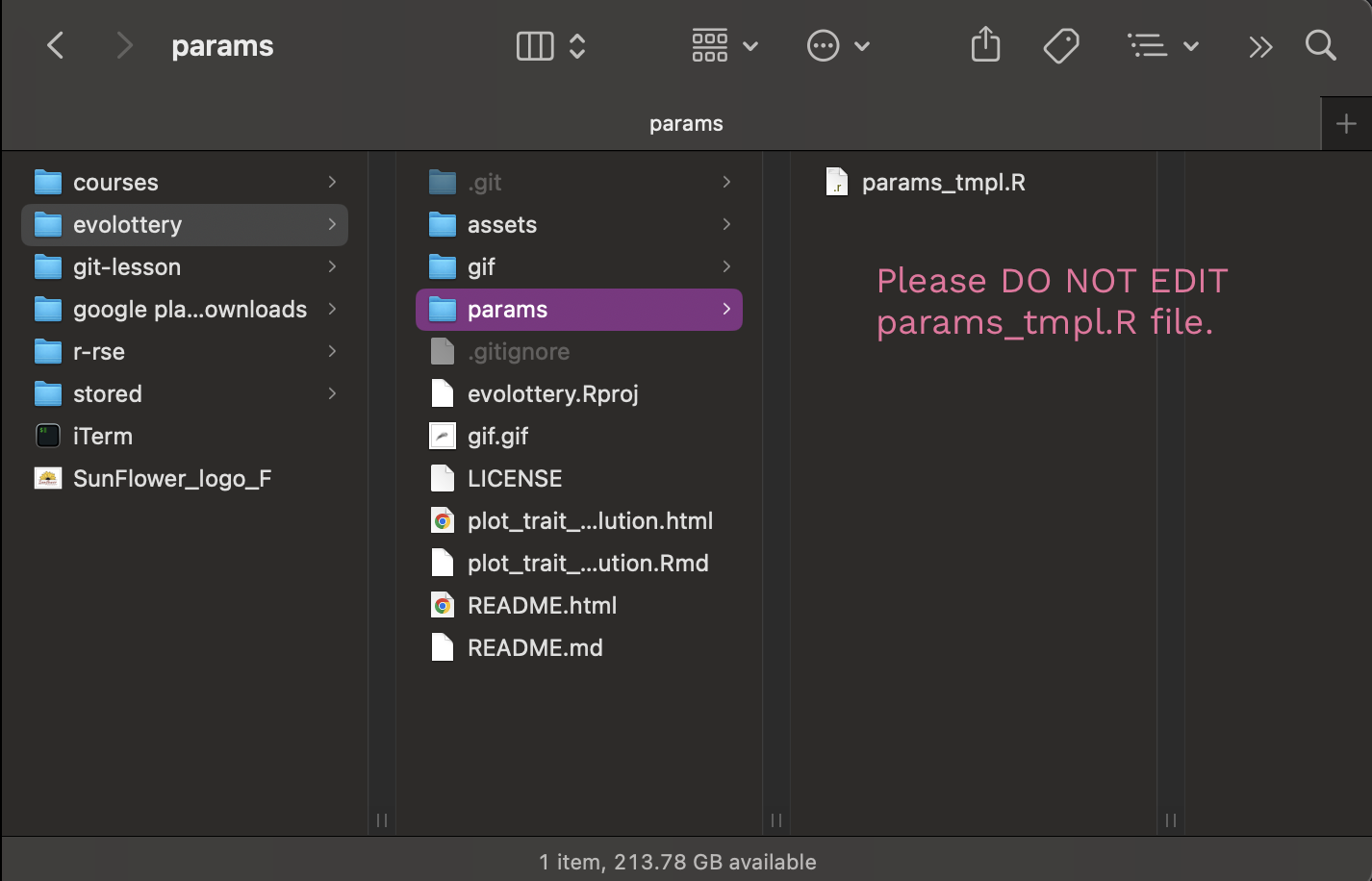
The params/ folder is where we are going to gather our individual parameter files. Currently, it just contains a .R template file called params_tmpl.R. Please DO NOT EDIT this file. We will make a copy of this file to edit.
Let’s go ahead and create and complete these files:
Open Repository in File System
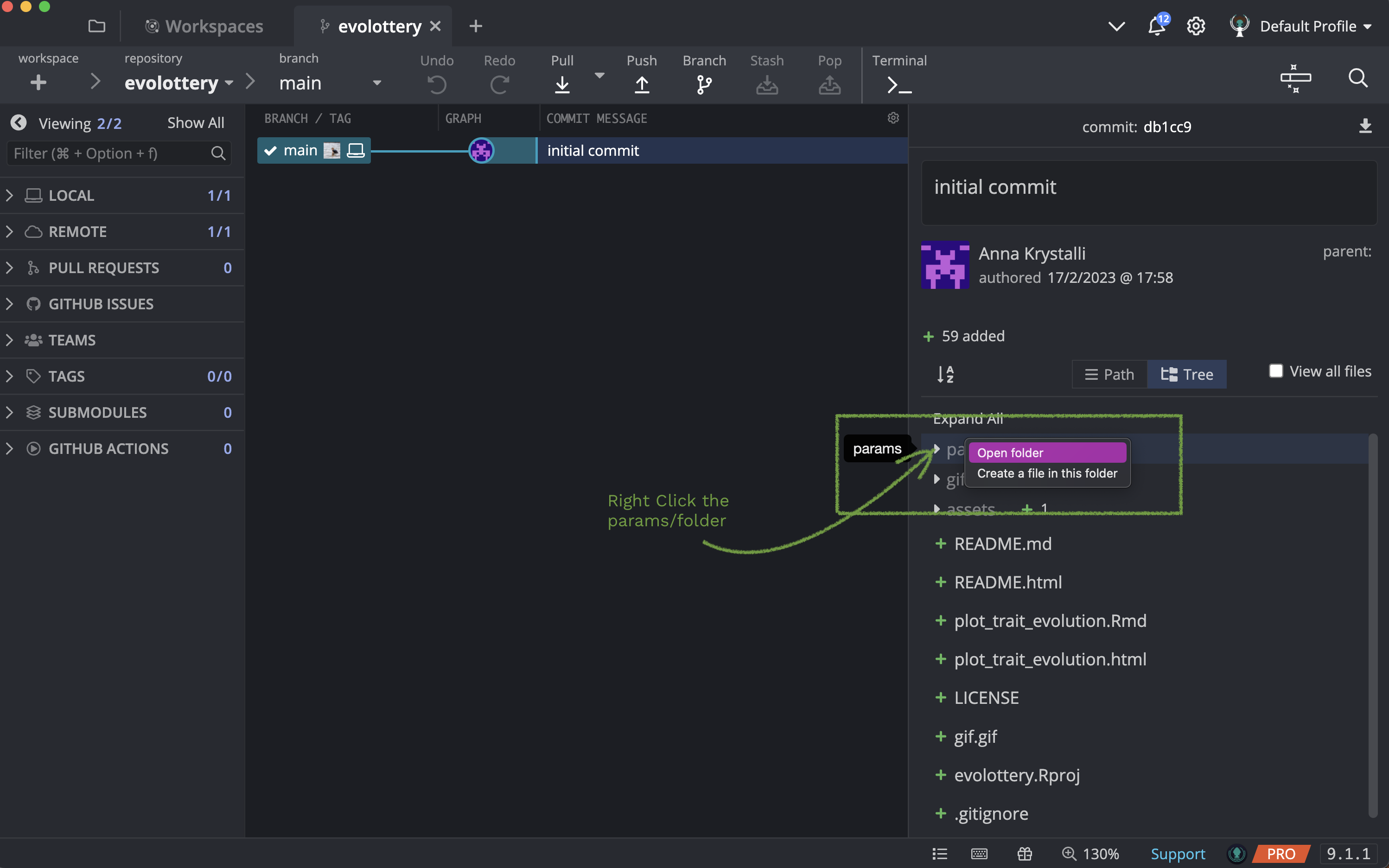
Right click on the params/ folder and choose to Open folder
You should now be looking at the contents of the params/ folder.
Currently, it just contains a .R template file called params_tmpl.R.
Please DO NOT EDIT this file. We will make a copy of this file to edit.
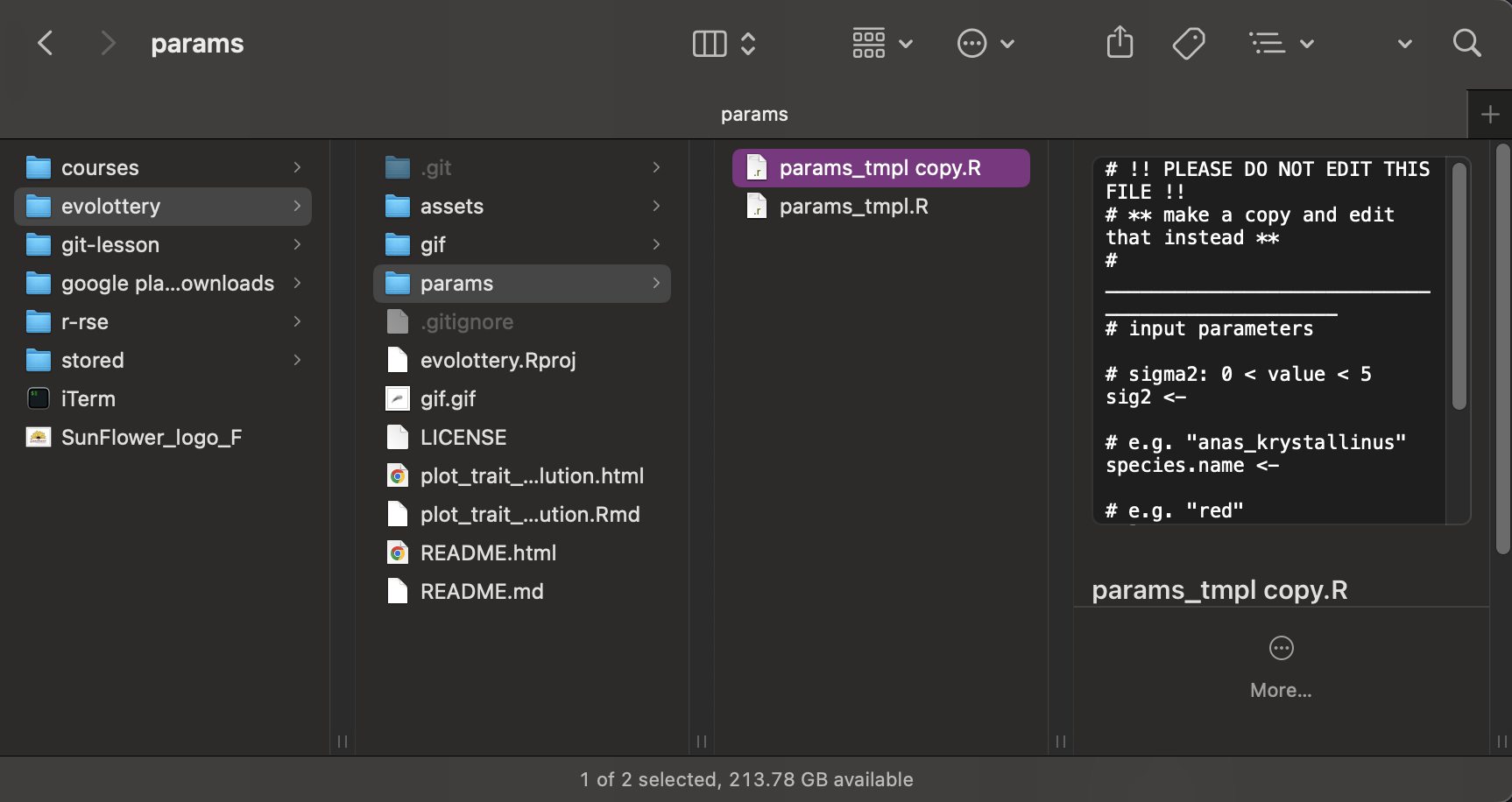
Make a copy of params/params_tmpl.R
First, make a copy of the file and save it in the same folder (params/).
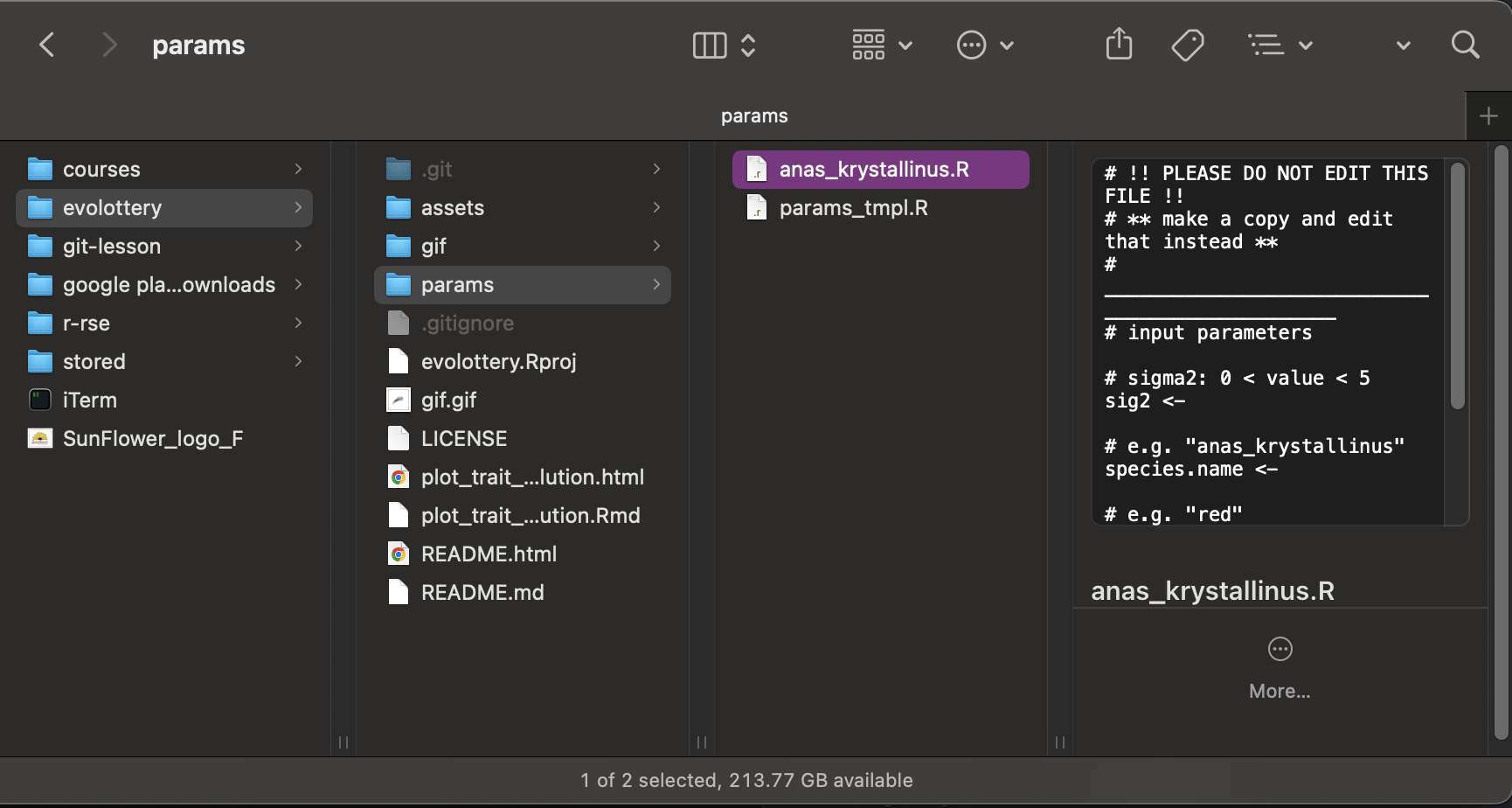
Now, think of a species name based on your name, eg Anas_krystallinus. This will be the species name associated with the evolutionary trajectory defined by the parameters you’re going to supply. This is both for for fun, but also to ensure that the files each participant commits has a unique name.
Use the species name you came up with to name the file.
Edit your parameter file
Let’s head back to GitKraken Client to edit the new file we created (although you could also do this in any text editor).
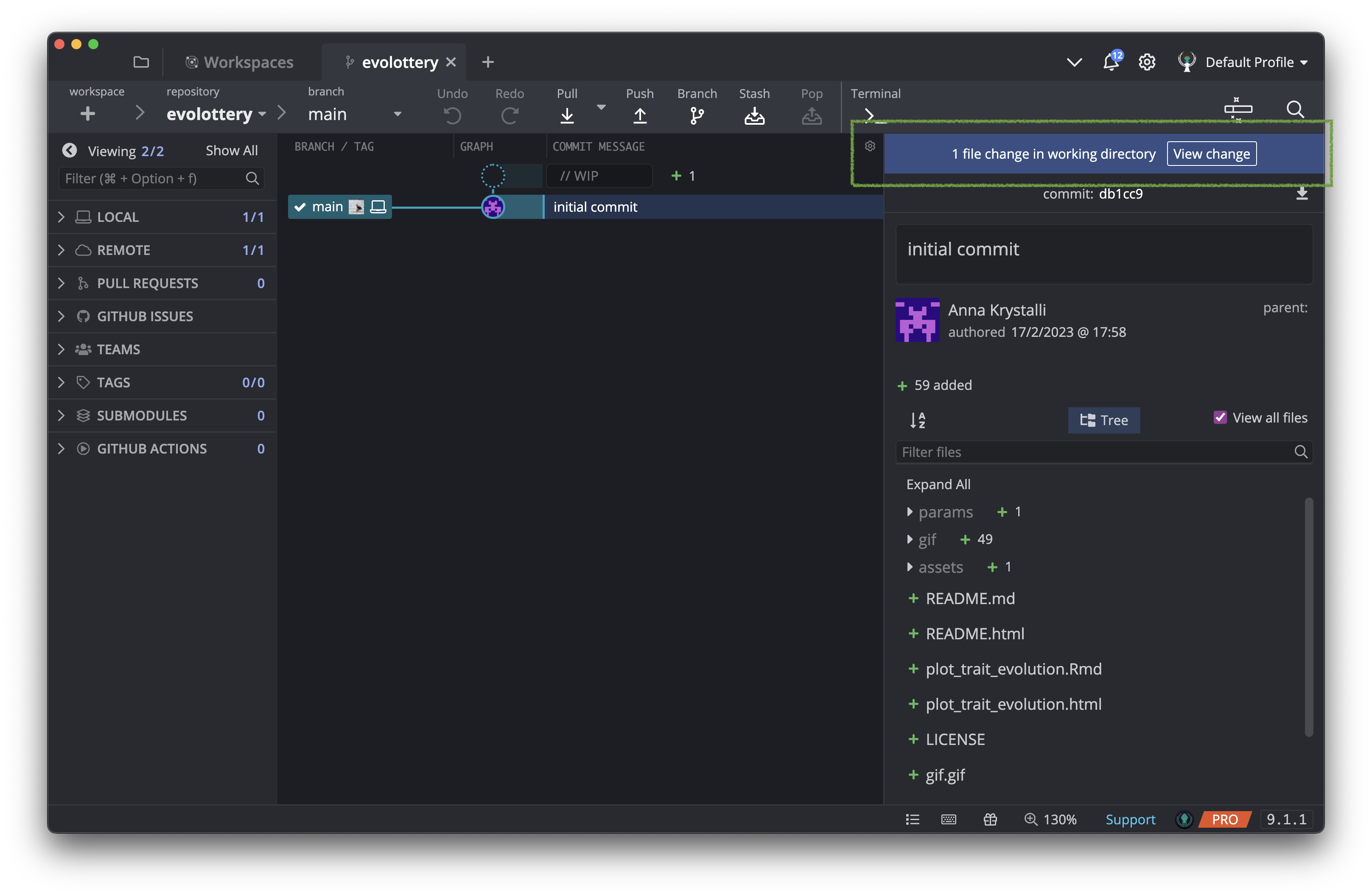
In the top of the right hand panel, GitKraken is telling us there’s been changes to a the repository. Click on View Changes.
Open your parameter file for editing
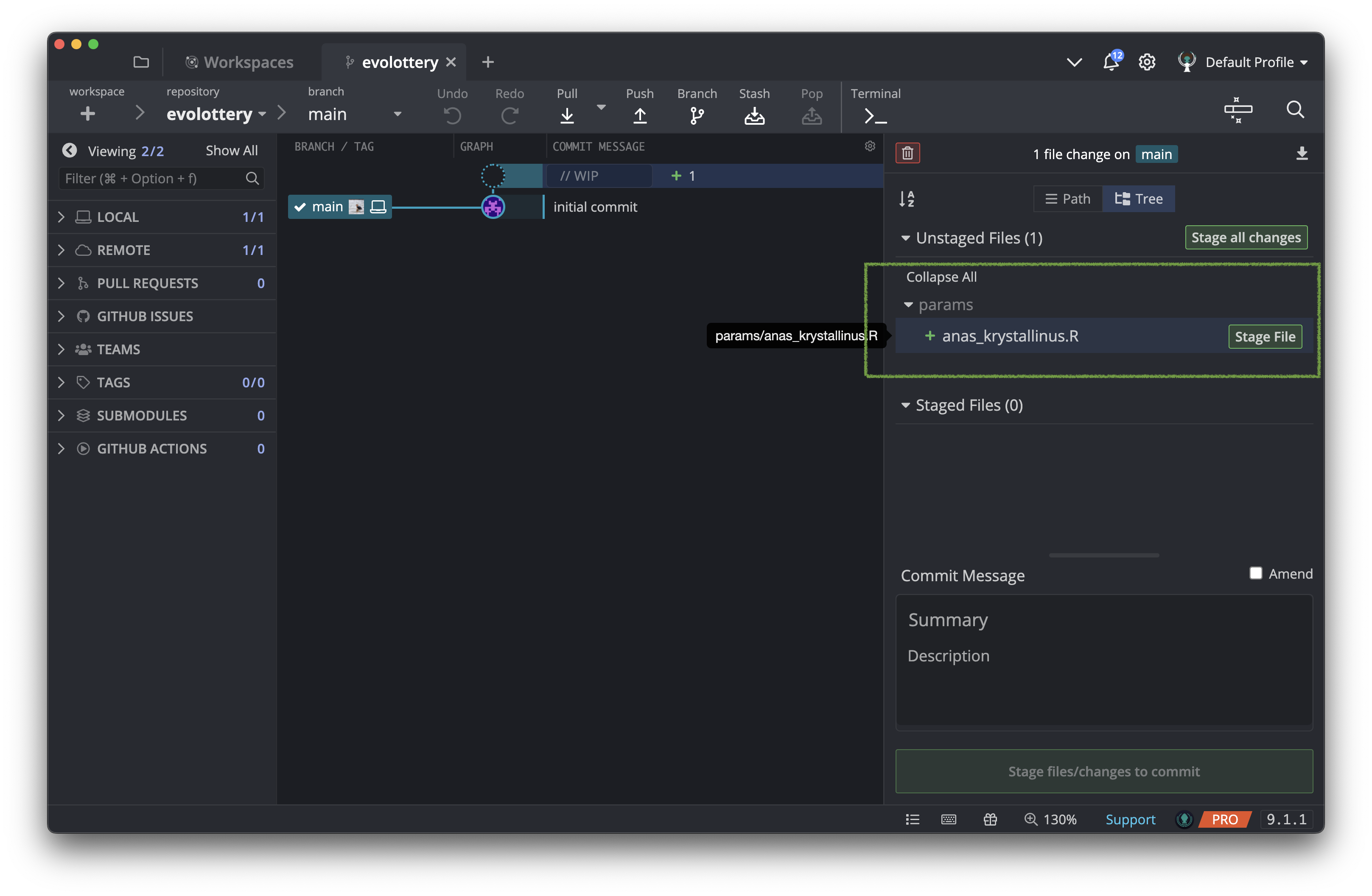
Now your parameter file is visible in the Unstaged Files panel.
Right click and choose to Edit file.
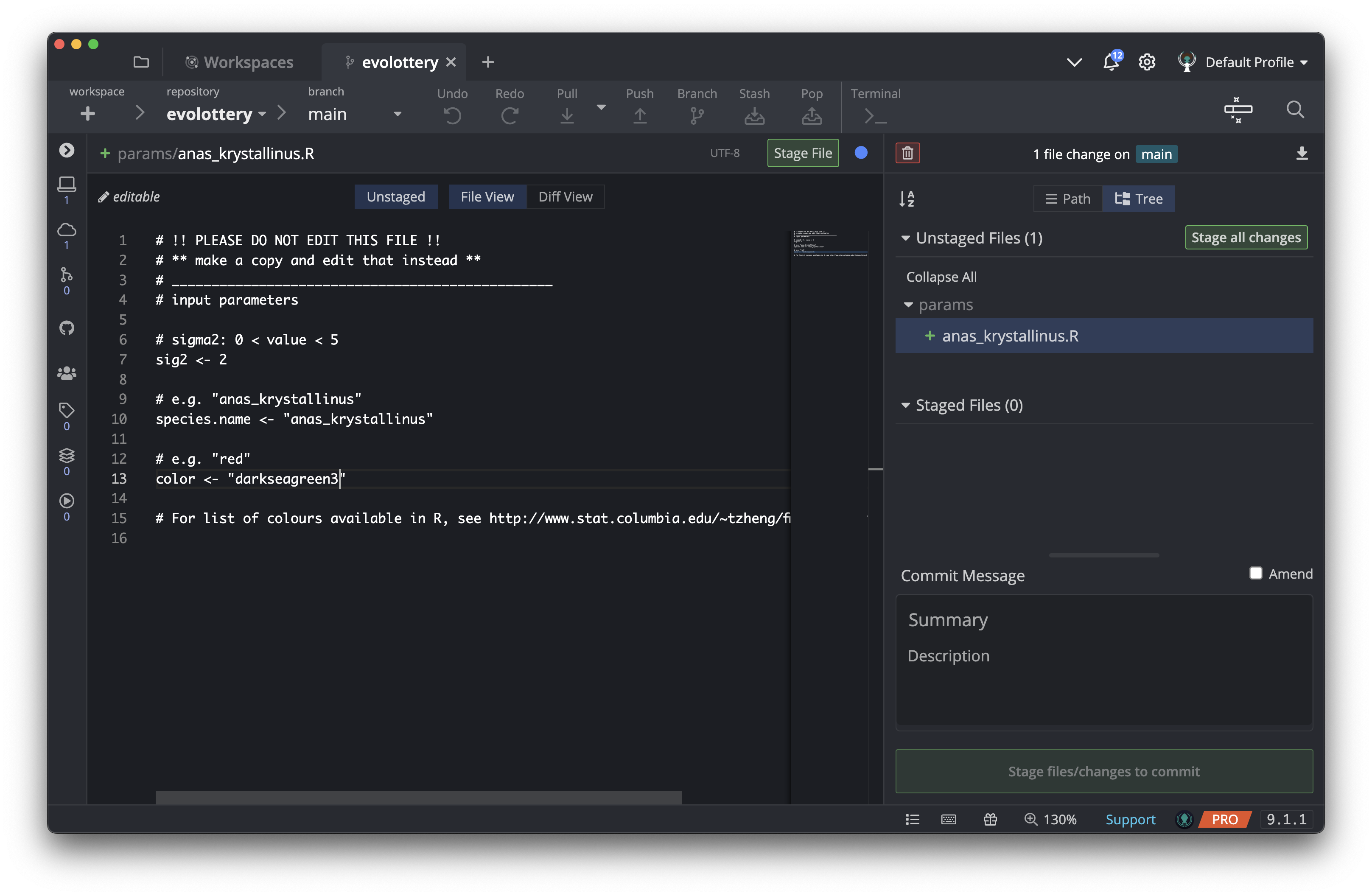
Edit your parameter file with values of your choice
Open the file you just created in your favourite text editor, edit it with parameters of your choice and save.
The parameters each participants need to supply are:
-
sig2: A numeric value greater than 0 but smaller than 5. -
species.name: a character string e.g."anas_krystallinus". Create a species name out of your name! It must be enclosed in double quotes (ie"...") -
color: a character string e.g."red","#FFFFFF"(Check out the list of available colours in R). It also must be enclosed in double quotes (ie"...")